NeuRA
The Neuron Reconstruction Algorithm
Reconstruct morphology from microscopy
Using raw microscopy data from, e.g. confocal or two-photon microscopes, NeuRA processes image stacks to automatically create surface reconstructions of underlying cells or organelles.
(Image by G. Queisser)

Anisotropic diffusion filtering
Image filters that analyze the local geometry using the inertial tensor allow guided filtering exclusively along structures. This enhances structure connectivity without distortion of the original geometry. The process of inertia-based, anisotropic filtering forms the basis of automatic geometry extraction.
(Image by G. Queisser)
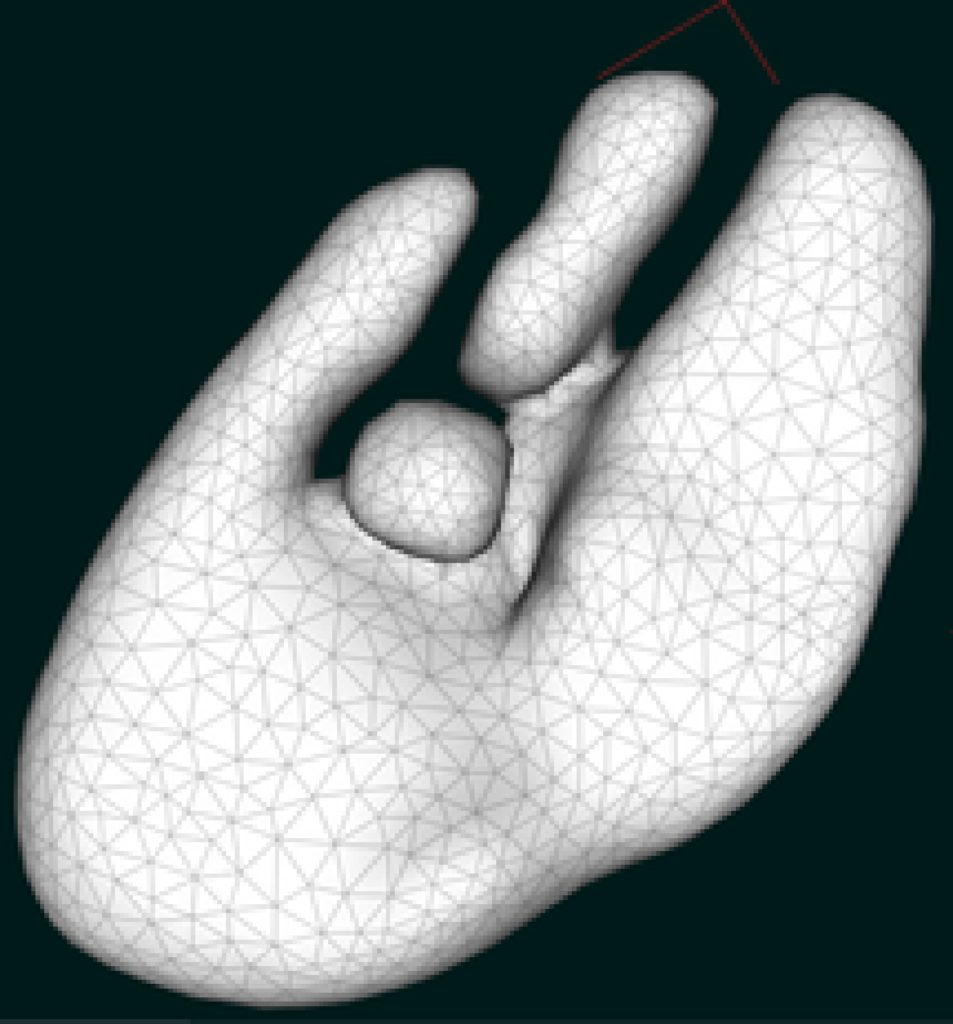
Surface and volume reconstruction
Running grid generating algorithms, like marching cubes, on filtered microscopy data yields a discrete surface representation (triangulation) of cells and organelles, from which discrete volume grids (e.g. tetrahedral grids) can be computed. The reconstructed computational domains can be directly integrated into detailed numerical simulations.
(Image by G. Queisser)
Contact us to get NeuRA
