
Numerics and Scientific Computing

Solving systems of PDEs
Having established a new direction in theory- and computation-driven biology, we are currently interested in advancing numerical methods and tools. Dealing with high-dimensional problems, we are now working on robust and physically coherent hybrid-dimensional numerical methods. Applying these to models, such as the Poisson-Nernst-Planck equations, we are able to demonstrate the possibility of reaching unprecedented biological accuracy through large scale computations. Our multiscale models and methods to translate molecular events to the functional level will push forward our numerics framework for detailed simulations in biology.
(Image by M. Stepniewski)
Major topics include
Partial Differential Equations
Ordinary Differential Equations
Discretization Methods
Fast Solvers, Multigrid Methods
Grid Generation from various sources
Software development
Some of our research projects in this area

Error estimation
Error estimators for non-linear PDEs with non-linear boundary conditions allow for adaptive grid refinement in grid-based numerical discretization. We develop estimators for equations that occur in biological and medical applications allowing us to solve very large problems on parallel computing architectures with good scaling properties.
(Image by M. Breit)
Hybrid discretization
Using surface and volume subdivision techniques in the context of multigrid grid refinement methods enables robust refinement with good regularity conditions. To guarantee global convergence, these refinement routines operate on octahedra/tetrahedra hybrid grids. Complex computational domains, often encountered in “real life” applications, can thus be incorporated in advanced numerical simulations.
(Image by M. Stepniewski)

Multiscale modeling and simulation
In biological processes multiple scales are often coupled and need to be considered when investigating a phenomenon on a single level. For instance, the molecular dynamics of receptor molecules can influence the function at a macroscopic level. Coupling the molecular dynamics locally with macroscopic, continuum dynamics is one of our focus projects and successfully applied in processes at cellular contact points, synapses.
(Image by S. Grein)
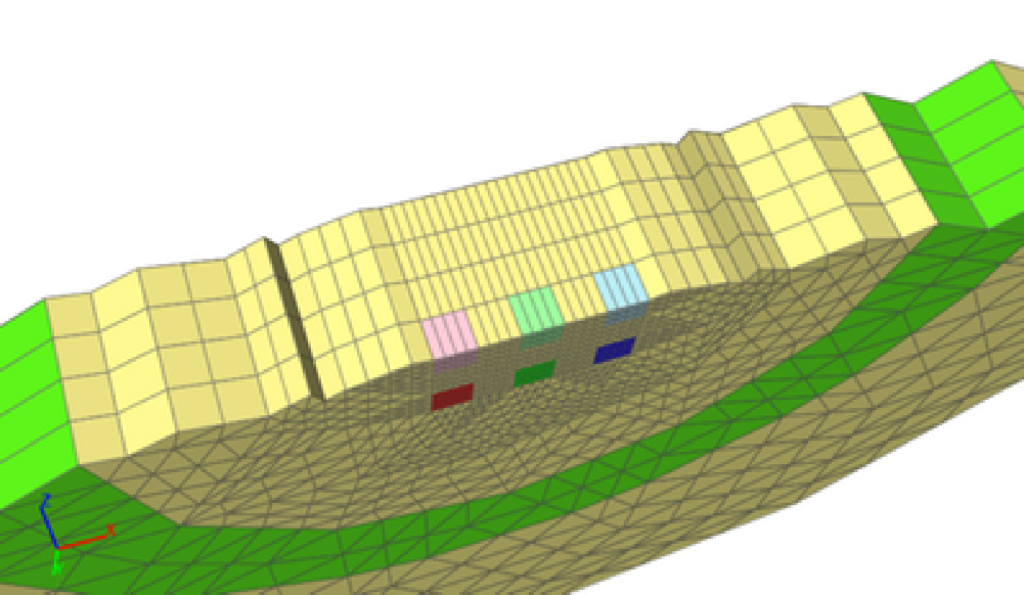
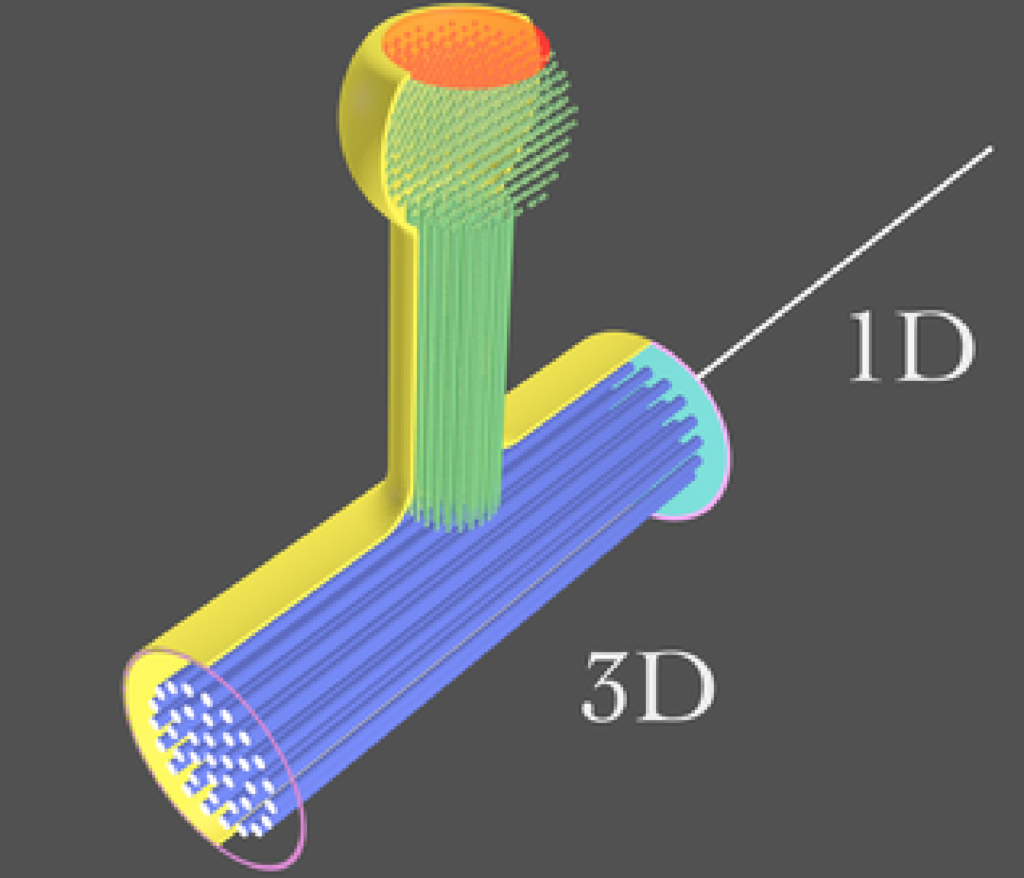
Hybrid-dimensional modeling and simulation
Computational complexity can restrict full resolution modeling and simulation to a subset of the actual problem size. Being able to couple high dimensional with low dimensional models is an essential component in addressing massive problem sizes. We are working on this topic in the context of the Poisson-Nernst-Planck equations and multi-dimensional neuron network modeling and simulation.
(Image by D. Patirniche)

Reconstruction methods
Including detailed geometry information in numerical computations can be quintessential in a given scientific context. We develop methods for automatic morphology reconstruction from microscopy data sets.
(Image by G. Queisser)
Multiphysics simulation framework
The simulation platform uG is being developed at the Goethe Center for Scientific Computing. Our group is a co-developer in the uG-project. Our numerical simulations are based on the uG-framework.

Neuroscientific simulations
We run a project termed NeuroBox which compiles all numerical tools and biological components in one toolbox. NeuroBox builds upon uG and the Visual Reflection Library (VRL) and its VRL-studio. Providing graphical workflow control allows non-experts to run complex numerical and neuroscientific computations.
(Image by G. Queisser)
